💊 AI should give us safer medicines
In-depth learning will give researchers better information about how a drug works in the human body.
Share this story!
Before a drug can be used, the manufacturer must know both that it has the right effect and what possible side effects it can cause. These tests require a lot of resources and take a lot of time. But now a research team has created a tool that can streamline work and also provide more information.
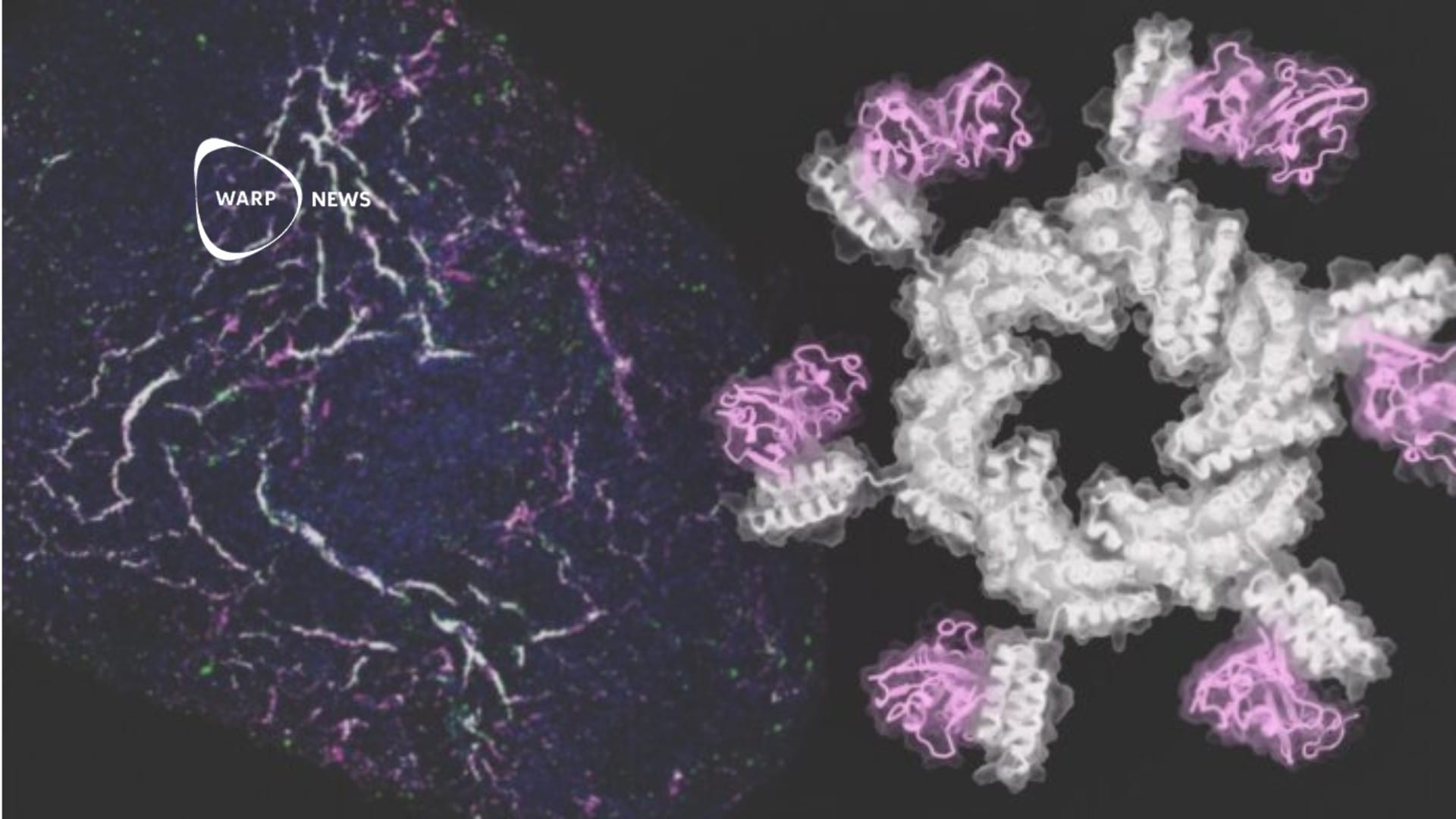
The research team from Rice University has used the in-depth AI technology to train a tool they call Metabolite Translator to predict which metabolites a drug may give rise to . A metabolite is a breakdown product that occurs when the molecules in the drug come in contact with enzymes in the body.
How safe a drug is depends not only on the drug but also on the metabolites that are created when the drug is activated in the body, says Eleni Litsa, lead author of the study that describes Metabolite Translator.
Normally, researchers examine enzymes in the liver to determine what effects a drug has had and whether it has given rise to any side effects. But metabolites can be created by other enzymes as well.
Our bodies are a network of chemical reactions. They have enzymes that react with chemicals and can break or create connections that change their structure to something that can be toxic or cause other complications. The existing methods focus on the liver because most non-copper substances are metabolized there. Instead, we try to describe human metabolism in general, says Eleni Litsa.
Eleni Litsa and the other researchers created Metabolite Translator using deep learning where the tool had to go through 900,000 known chemical reactions. Based on these data, Metabolism Translator learned to predict which metabolites different reactions can cause.
Less manual work
There are already other tools that can predict which metabolites a drug can give rise to. But these tools are based on experts manually entering the rules to be followed. Which of course is labor intensive and expensive, plus the tools can not detect something that is not covered by a rule.
To see how well Metabolite Translator works, the researchers compared it with today's tools. The result was that Metabolite Translator found the same enzymes as the traditional methods, but that it also identified enzymes that are not normally involved in metabolizing drugs, but which nevertheless gave rise to metabolites and which the traditional methods missed.
We have a system that works as well as rule-based systems and we did not use predefined rules, which require manual work and expert knowledge, in our system. This would not even have been possible just two years ago, says Lydia Kavraki, professor of biotechnology at Rice University and co-author of the study.
With Metabolite Translator, researchers and pharmaceutical companies can now get a tool that can better describe how a drug affects the whole body. It also does not have to rely on experts spending their valuable time entering rules into a database, but can instead devote themselves to more advanced tasks.
By becoming a premium supporter, you help in the creation and sharing of fact-based optimistic news all over the world.